SARS-CoV-2 Publications
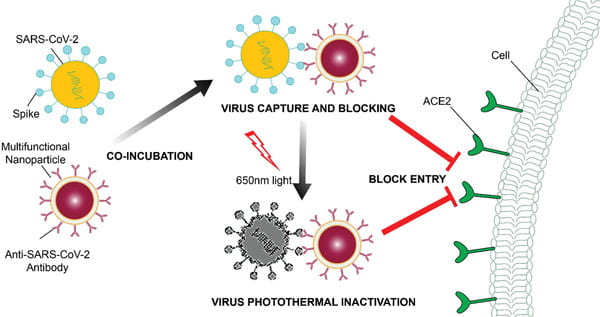
Cai X, Chen M, Prominski A, Lin Y, Ankenbruck N, Rosenberg J, Nguyen M, Shi J, Tomatsidou A, Randall G, Missiakas D, Fung J, Chang EB, Penaloza-MacMaster P, Tian B, Huang J. A Multifunctional Neutralizing Antibody-Conjugated Nanoparticle Inhibits and Inactivates SARS-CoV-2. Adv Sci (Weinh). 2022 Jan;9(2):e2103240. doi: 10.1002/advs.202103240. Epub 2021 Nov 10. PMID: 34761549; PMCID: PMC8646742.
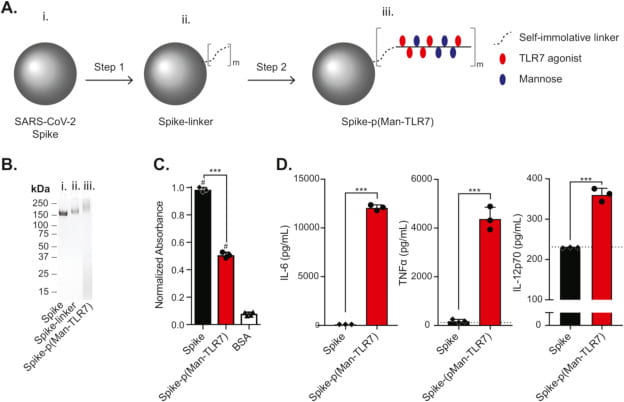
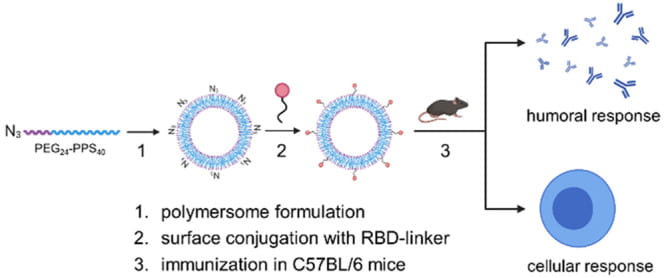
Gray LT, Raczy MM, Briquez PS, Marchell TM, Alpar AT, Wallace RP, Volpatti LR, Sasso MS, Cao S, Nguyen M, Mansurov A, Budina E, Watkins EA, Solanki A, Mitrousis N, Reda JW, Yu SS, Tremain AC, Wang R, Nicolaescu V, Furlong K, Dvorkin S, Manicassamy B, Randall G, Wilson DS, Kwissa M, Swartz MA, Hubbell JA. Generation of potent cellular and humoral immunity against SARS-CoV-2 antigens via conjugation to a polymeric glyco-adjuvant. Biomaterials. 2021 Nov;278:121159. doi: 10.1016/j.biomaterials.2021.121159. Epub 2021 Sep 30. PMID: 34634664; PMCID: PMC8482845.
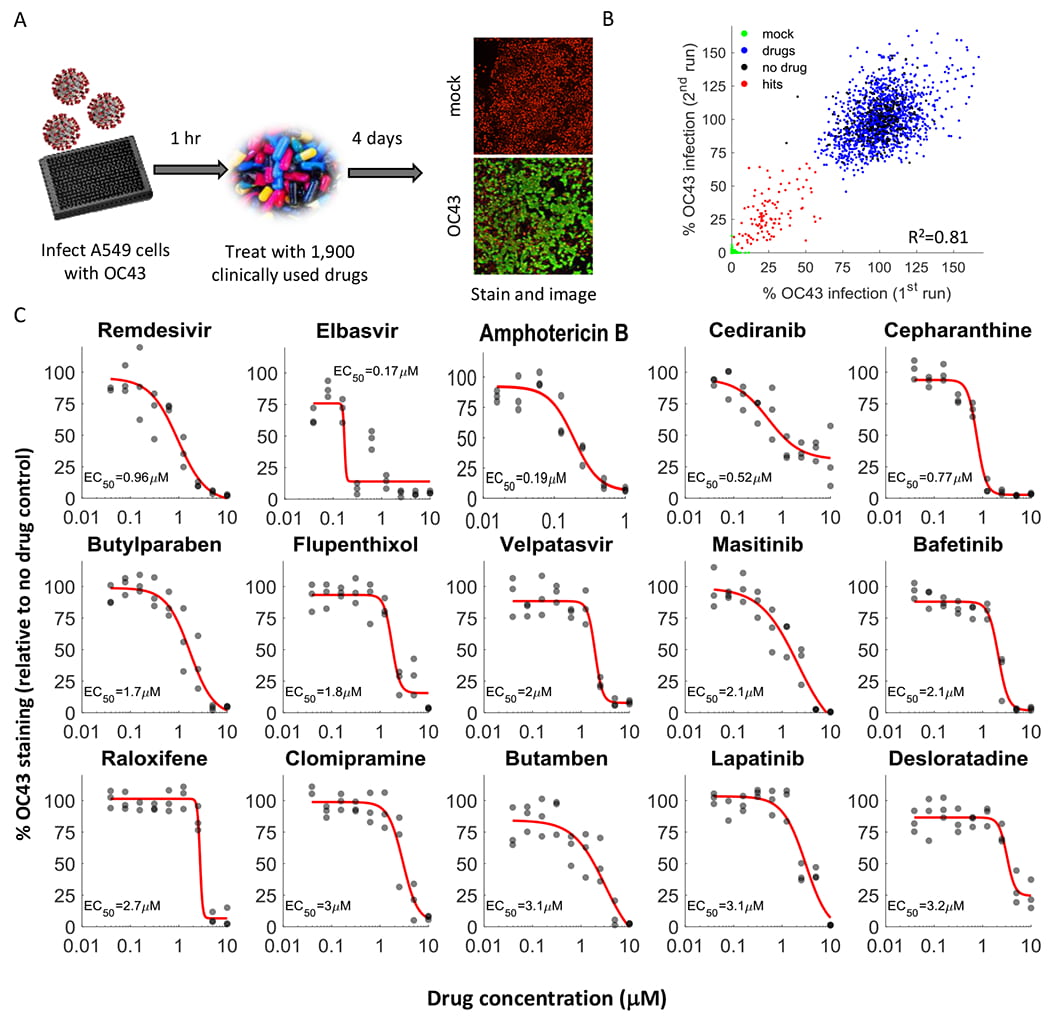
Drayman N, DeMarco JK, Jones KA, Azizi SA, Froggatt HM, Tan K, Maltseva NI, Chen S, Nicolaescu V, Dvorkin S, Furlong K, Kathayat RS, Firpo MR, Mastrodomenico V, Bruce EA, Schmidt MM, Jedrzejczak R, Muñoz-Alía MÁ, Schuster B, Nair V, Han KY, O’Brien A, Tomatsidou A, Meyer B, Vignuzzi M, Missiakas D, Botten JW, Brooke CB, Lee H, Baker SC, Mounce BC, Heaton NS, Severson WE, Palmer KE, Dickinson BC, Joachimiak A, Randall G, Tay S. Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2. Science. 2021 Aug 20;373(6557):931-936. doi: 10.1126/science.abg5827. Epub 2021 Jul 20. PMID: 34285133; PMCID: PMC8809056.
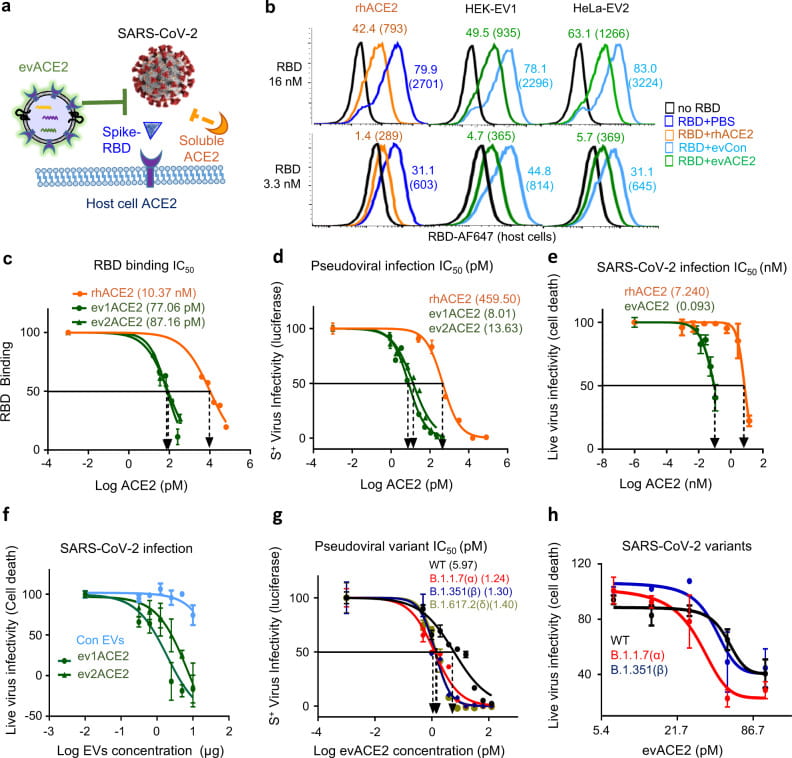
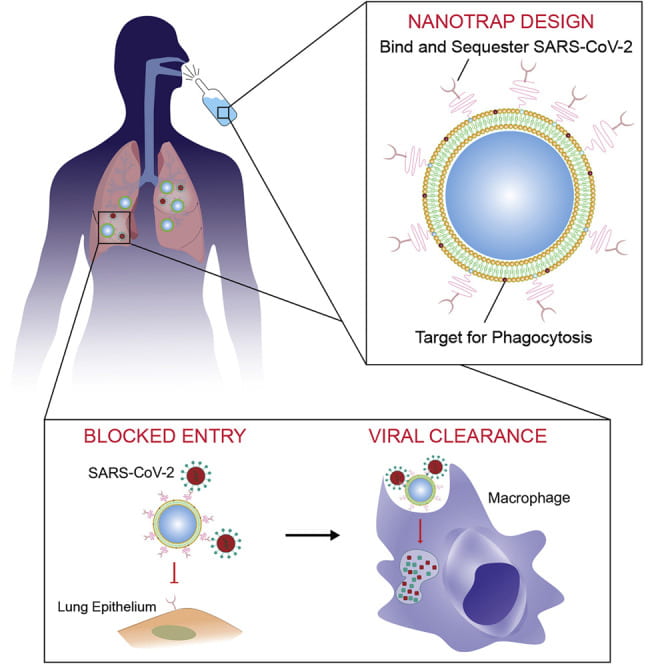
Chen M, Rosenberg J, Cai X, Hsuan Lee AC, Shi J, Nguyen M, Wignakumar T, Mirle V, Edobor AJ, Fung J, Donington JS, Shanmugarajah K, Lin Y, Chang E, Randall G, Penaloza-MacMaster P, Tian B, Madariaga ML, Huang J. Nanotraps for the containment and clearance of SARS-CoV-2. Matter. 2021 Jun 2;4(6):2059-2082. doi: 10.1016/j.matt.2021.04.005. Epub 2021 Apr 22. PMID: 33907732; PMCID: PMC8062026.
Teplensky MH, Distler ME, Kusmierz CD, Evangelopoulos M, Gula H, Elli D, Tomatsidou A, Nicolaescu V, Gelarden I, Yeldandi A, Batlle D, Missiakas D, Mirkin CA. Spherical nucleic acids as an infectious disease vaccine platform. Proc Natl Acad Sci U S A. 2022 Apr 5;119(14):e2119093119. doi: 10.1073/pnas.2119093119. Epub 2022 Mar 21. PMID: 35312341.
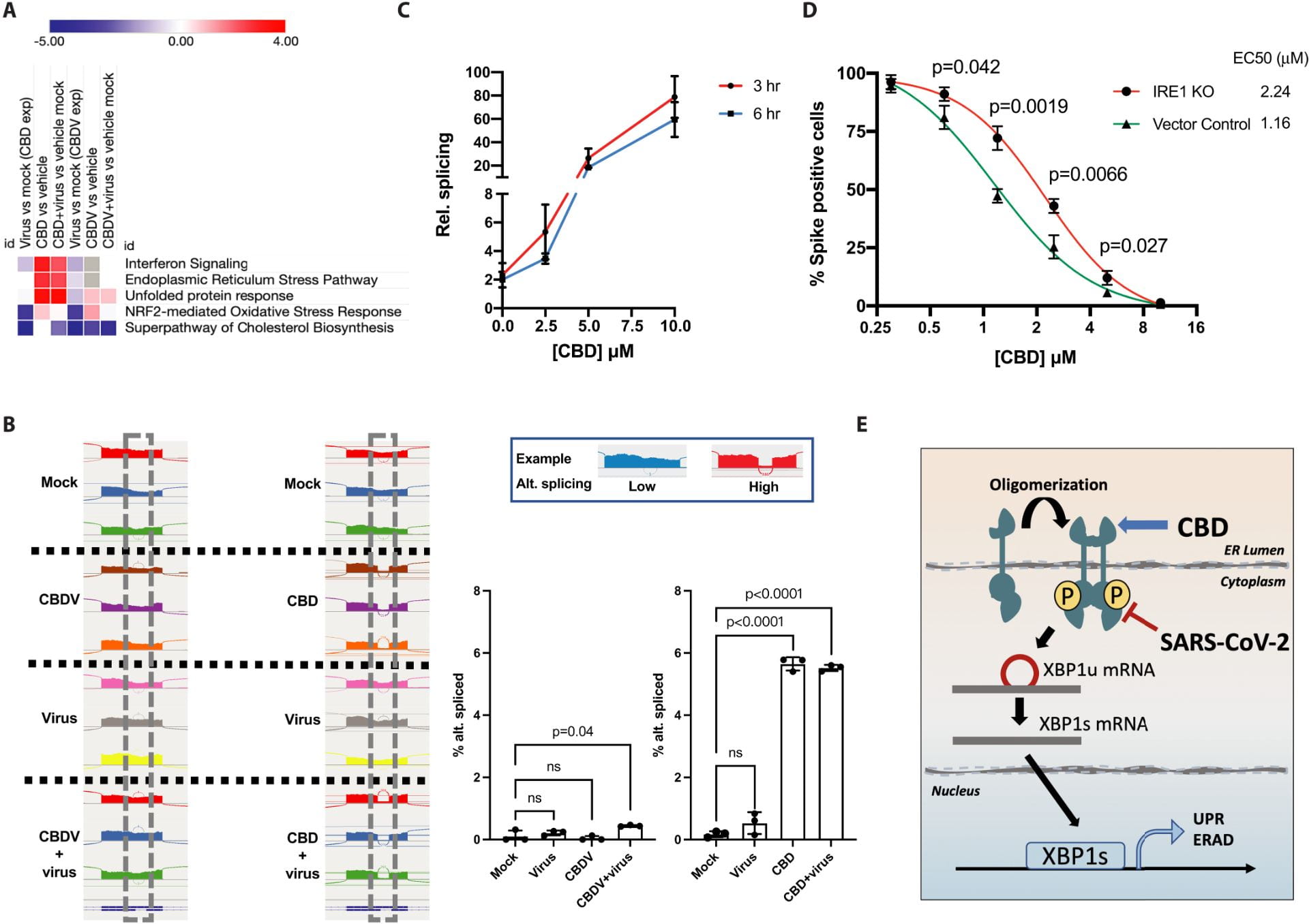
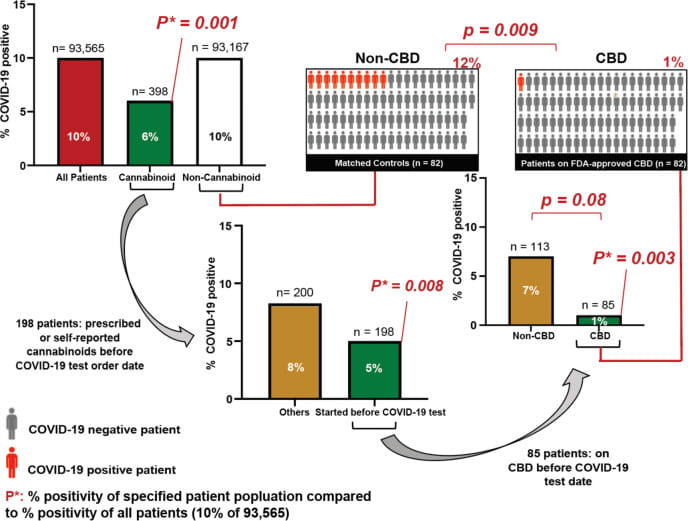
Nguyen LC, Yang D, Nicolaescu V, Best TJ, Ohtsuki T, Chen SN, Friesen JB, Drayman N, Mohamed A, Dann C, Silva D, Gula H, Jones KA, Millis JM, Dickinson BC, Tay S, Oakes SA, Pauli GF, Meltzer DO, Randall G, Rosner MR. Cannabidiol Inhibits SARS-CoV-2 Replication and Promotes the Host Innate Immune Response. bioRxiv [Preprint]. 2021 Mar 10:2021.03.10.432967. doi: 10.1101/2021.03.10.432967. PMID: 33758843; PMCID: PMC7987002.